
|
電子情報通信学会 - IEICE会誌 試し読みサイト
© Copyright IEICE. All rights reserved.
|

|
電子情報通信学会 - IEICE会誌 試し読みサイト
© Copyright IEICE. All rights reserved.
|

Abstract
Smart farming is on the uprising demand all over the world. Artificial Intelligence (AI) is considered as one of the latest tools to be utilized for smart farming. However, the practical implementation of AI for smart farming is often a challenge. One of the challenges is to optimize the algorithms for accurate classification of plant diseases. In this study, we have proposed a Convolutional Neural Network (CNN) for the classification of leaf diseases. The framework of the proposed CNN is designed using the Depthwise Separable Convolution (DWS) technique that consists of two stages, i. e., depthwise and pointwise feature extractions. We have compared the model with the classical convolutional approach. Results show that the proposed model outperformed the conventional CNN model with a precision of 0.932, recall 0.992, F1 score of 0.961 and a test accuracy of 95.25% whereas the conventional model achieved precision 0.941, recall 0.961, F1 score 0.951 and 93.76% of test accuracy.
Keywords:leaf diseases, image classification, convolutional neural networks
Finding sustainable methods to generate enough food to feed everyone is crucial because of the world’s continually growing population. Innovative solutions for the agricultural sector, including smart farming methods to boost productivity and efficiency, have been made possible by modern technology(1)~(3). Smart farming involves the use of cutting-edge technology to improve a variety of farming operations, including irrigation, fertilization, and pest control, such as Internet of Things (IoT) devices, drones, and machine learning algorithms(4),(5). But despite these developments, a major problem, food security, still exists. According to the Food and Agriculture Organization (FAO), plant diseases are thought to be responsible for a sizable share of yearly crop losses that result in billions of dollars in economic losses(6). Unpredictable weather patterns, land degradation, and population increase make the issue worse. Finding strategies to reduce crop losses and preserve global food security is crucial(3),(7).
Fortunately, scientists and researchers are actively seeking solutions to these problems. This includes the development of new crop varieties that are resistant to diseases and pests, as well as the use of innovative technologies to identify and diagnose plant diseases in their early stages(8). Additionally, precise farming techniques that enable farmers to make data-driven decisions about their crops can help reduce losses and increase yields(9).
Global food security is a complicated, multifaceted subject that necessitates creative solutions(10). Smart farming technologies have been made possible by contemporary technology, but more research and innovation are urgently needed to provide a sustainable and reliable food supply for the world’s expanding population(4),(11).
Many scientists have worked hard to create efficient techniques for spotting and stopping plant diseases in their early stages. Early identification is essential because it enables farmers to act right away to stop the disease’s spread and save crop losses.
To achieve this goal, many researchers have turned to computer vision-based techniques, including machine learning and deep learning(4),(11). These methods involve training algorithms to recognize patterns and identify the early signs of disease in plant images. By analyzing these images, these algorithms can detect changes in plant color, texture, and shape, which may indicate the presence of a disease(4).
The traditional methods of leaf classification relied heavily on manual feature extraction, which was time-consuming, subjective, and limited in terms of capturing the intricate details and patterns present in leaf images. In contrast, deep learning techniques like CNNs have shown great potential in automatically learning discriminative features directly from raw data, eliminating the need for manual feature engineering. LeafNet(12) was proposed as a CNN model for leaf classification, aiming to overcome the limitations of manual feature extraction in automated plant species identification. By using CNNs, LeafNet could learn discriminative features directly from raw leaf images, enabling accurate and efficient leaf recognition. It achieved high accuracy rates and outperformed traditional methods. However, researchers sought to improve LeafNet further by incorporating techniques like depthwise and pointwise convolutions, which aimed to enhance efficiency without sacrificing accuracy.
This study aimed to improve the accuracy of the LeafNet model(12) for classifying plant diseases by modifying its architecture. The original LeafNet model was designed using a sequential convolutional layer approach, which has some limitations. Sequential models rely on a fixed order of operations, which may not effectively capture complex patterns and relationships within the input data. They may struggle to handle spatial dependencies and can result in limited representational capacity.
To address these limitations, we introduced depthwise and pointwise convolutional layers(13) into the LeafNet architecture. Depthwise convolutional layers perform spatial filtering independently for each input channel, capturing more fine-grained details and allowing the model to better understand local patterns within the image. Pointwise convolutional layers further enhance the model’s ability to capture higher-level features by projecting the output of depthwise convolutions into a new feature space. By incorporating these layers, we aim to enhance the model’s capability to extract meaningful and discriminative features from the input data, thereby improving classification accuracy and enabling better generalization to unseen rice leaf disease samples.
By doing so, we were able to reduce the number of parameters required for the model while still improving its accuracy compared to the original LeafNet model.
The addition of depthwise and separable convolutional layers allowed the model to more efficiently extract and process features from the input data, resulting in a more accurate classification of plant diseases. This approach also helped to reduce the risk of overfitting the model to the training data, making it more robust and reliable in real-world applications.
This paper comprises of 5 sections, Background study is given in Section 2, whereas Section 3 describes about the methodology adopted in this study. Section 4 presents the results and discussions, and Section 5 summarizes and concludes this study.
Researchers interest in automating the detection and classification of plant diseases has grown during the past several years. Utilizing CNNs is one strategy that has shown promise in this field. CNNs are a class of deep learning models that have gained significant popularity in computer vision tasks, including the classification of diseases. CNNs are specifically designed to effectively process and analyze visual data, making them well-suited for image-based tasks.
CNNs have been used for a variety of tasks, such as disease diagnosis, crop monitoring, intelligent spraying, yield and price prediction, crop and soil monitoring, disease diagnosis, and farm monitoring(14).
The key characteristic of CNNs lies in their ability to automatically learn hierarchical representations from raw image data. Unlike traditional machine learning algorithms, CNNs can automatically extract relevant features from images through a series of convolutional and pooling layers. These layers perform local receptive field operations, allowing the network to capture spatial relationships and patterns in the input images.
The use of CNNs for plant disease classification is particularly promising because they are well-suited to processing large amounts of image data, such as the images of diseased that are used for diagnosis(15),(16). By training the CNN on a large dataset of labeled images, it can learn to automatically identify patterns and features that are indicative of specific diseases, making it possible to classify new images with a high degree of accuracy.
Gokulnath et. al.(17) proposed a CNN model for disease identification on the Plant Village dataset. Their model utilized a loss-fusion technique that combined multiple loss functions to improve the accuracy of the model. The proposed model achieved an impressive accuracy of 98.93%, demonstrating the potential of CNNs for accurately identifying plant diseases. This study highlights the importance of utilizing advanced machine learning techniques to improve the accuracy of disease identification models, which can ultimately lead to more effective disease management strategies in agriculture.
Chen et. al.(18) proposed an improvement on the final layer of the VGG model, which was originally trained on the ImageNet dataset. They transferred the learned feature information to the Inception Module and applied it to a merged dataset of maize and rice. The proposed model achieved an accuracy of 92%, indicating the effectiveness of the transfer learning approach. This study highlights the importance of utilizing pre-trained models and transfer learning techniques to improve the accuracy of plant disease classification models. By building upon the knowledge acquired from previous models, researchers can develop more accurate and efficient solutions for identifying plant diseases, which can have significant benefits for agriculture and food security.
Shijie et. al.(19) proposed a methodology for tomato disease classification using the VGG16 model combined with the Multi-Class Support Vector Machine (MSVM) algorithm. Their study involved the classification of ten different disease classes, and they achieved an accuracy of 89%. This study highlights the potential of combining deep learning models with traditional machine learning algorithms to improve the accuracy of plant disease classification. Sahu et al.(20) conducted an experiment on two popular CNN models, namely GoogleNet and VGG16, to classify beans in a dataset. The results showed that GoogleNet outperformed VGG16, achieving an accuracy of 95%. This study demonstrated the effectiveness of CNN models in accurately classifying different types of beans, which can have significant implications for improving crop yield and food security. The findings suggest that GoogleNet may be a suitable model for similar classification tasks involving plant diseases or other agricultural applications.
Tiwari et. al.(21) proposed a novel deep convolutional neural network to identify 27 diseases in six different crops. They enhanced the CNN on a complicated and difficult dataset, achieving an impressive cross-validation accuracy of 99.58% on average. This study highlights the potential of deep learning techniques for accurate and efficient detection of multiple plant diseases, which can ultimately aid in the improvement of crop yield and food security.
Pierre Barré et. al.(12) introduced a novel CNN called LeafNet, which was developed to classify plant leaves based on their level of infection. The authors reported achieving high accuracy levels of 98.69% and 98.75% on smaller datasets, while they achieved a lower accuracy of 79.66% on a larger dataset. This study provided an initial framework for further improvements in the detection and classification of plant diseases using deep learning techniques. Similarly, this study aimed to improve the accuracy of the LeafNet model for classifying plant diseases by modifying its architecture. To achieve this, we added depthwise and separable convolutional layers to the base model. By doing so, we were able to reduce the number of parameters required for the model while still improving its accuracy compared to the original LeafNet model. The addition of depthwise and separable convolutional layers allowed the model to more efficiently extract and process features from the input data, resulting in a more accurate classification of plant diseases. This approach also helped to reduce the risk of overfitting the model to the training data, making it more robust and reliable in real-world applications.
Our modified architecture provided a more efficient and accurate solution for classifying plant diseases, with reduced computational requirements compared to the original LeafNet model. This approach has the potential to improve the efficiency and effectiveness of disease diagnosis in the agricultural industry.
We utilized the New Plant Disease Dataset(22)which consists of 87,000 images of diseased leaves across 38 categories. Out of these, we used 70,295 images for training and validation while 16,705 images were reserved for testing purposes. To enhance our model performance, we split the training dataset into a ratio of 90: 10 for training and validation, respectively. As a result, our training dataset contains 63,282 images while the validation dataset contains 7,013 images, both distributed across 38 classes. Fig.1 shows the sample images from the dataset with their respective disease. The New Plant Disease Dataset available on Kaggle(22)comprises 38 categories of leaf diseases including Apple, Potato, Raspberry, Strawberry, Squash, Corn, Grape, Blueberry, Soybean, Peach, and Cherry.

In order to prepare the dataset for training, we applied various pre-processing techniques on the images. Firstly, we resized all images to a fixed size of 224×224 pixels. This resizing was done to ensure consistency in image sizes across the dataset. Next, we performed data augmentation techniques to increase the diversity of the dataset and to prevent overfitting of the model. Data augmentation techniques include rotation, flipping, and shifting of the images. After data augmentation, we normalized the pixel values of the images to ensure that they were within a range of 0 to 1. Overall, the pre-processing and augmentation steps were crucial in ensuring that the dataset was diverse and suitable for training the deep learning model. These steps helped to improve the accuracy of the model and prevent overfitting, resulting in better generalization performance.
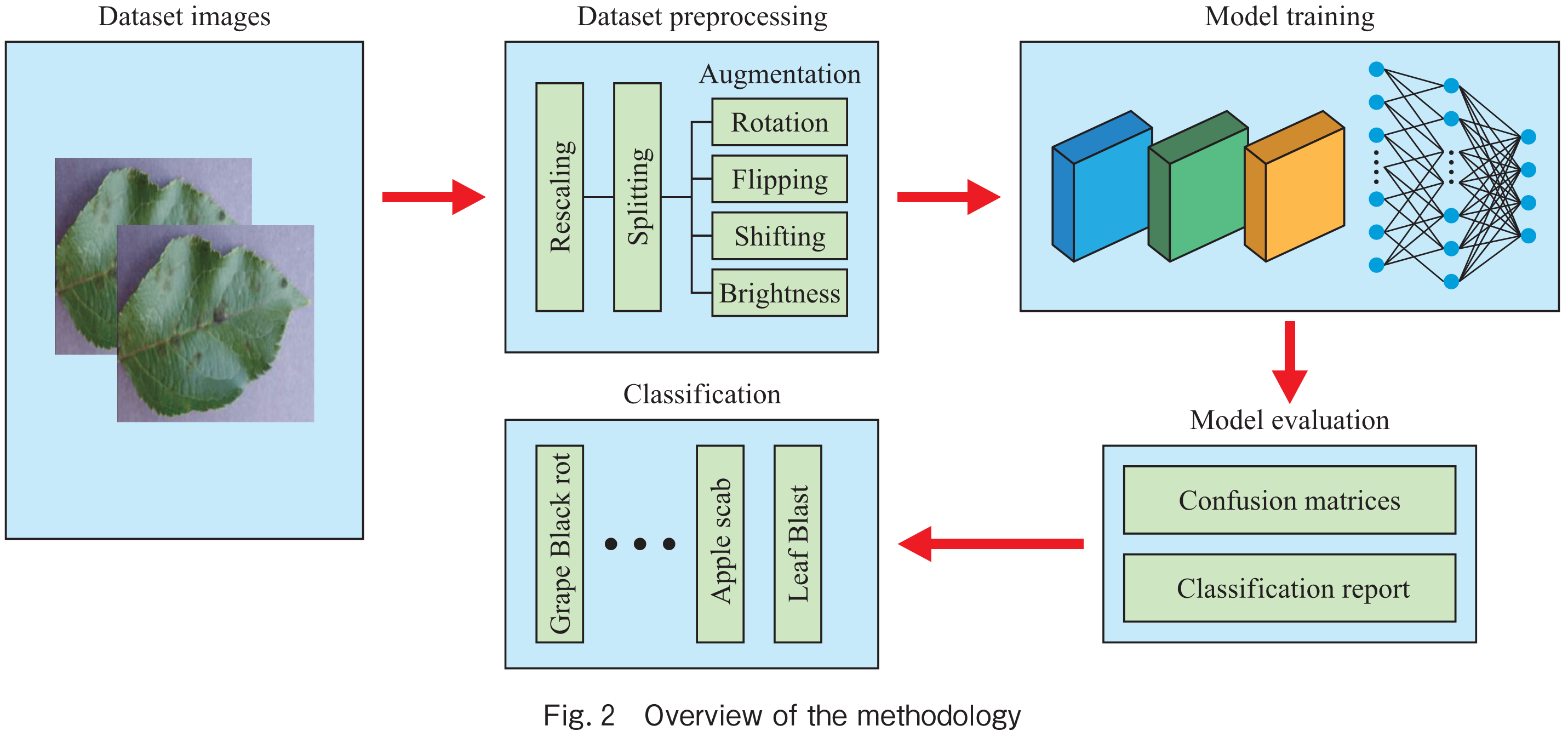
The present study employed a specific methodology, which is summarized in Fig.2. The figure illustrates various components, including dataset images, data pre-processing, model training, model evaluation, and classification. First step is of collection of dataset images, the information about the dataset images has already been described in section 3.1. Similarly, the second step in our proposed methodology is data preprocessing and data augmentation, for data preprocessing we used various techniques like resizing, normalization and converting the categorical labels into one-hot encoding. For data augmentation to make the dataset more diverse, we applied random rotations, horizontal and vertical flips, and brightness and contrast adjustments to the images.
For the model training, we used LeafNet as a base model and modified it by adding Depthwise and Separable Convolutional Layers to reduce the number of parameters and improve the accuracy of the model. The modified model was trained on the pre-processed and augmented dataset.
For model evaluation, we evaluated the performance of our model using various metrics such as accuracy, precision, recall, and F1 score. We also used confusion matrix to analyze the classification results.
The final step i.e., classification, we used our trained model to classify the test set images into their respective disease categories.
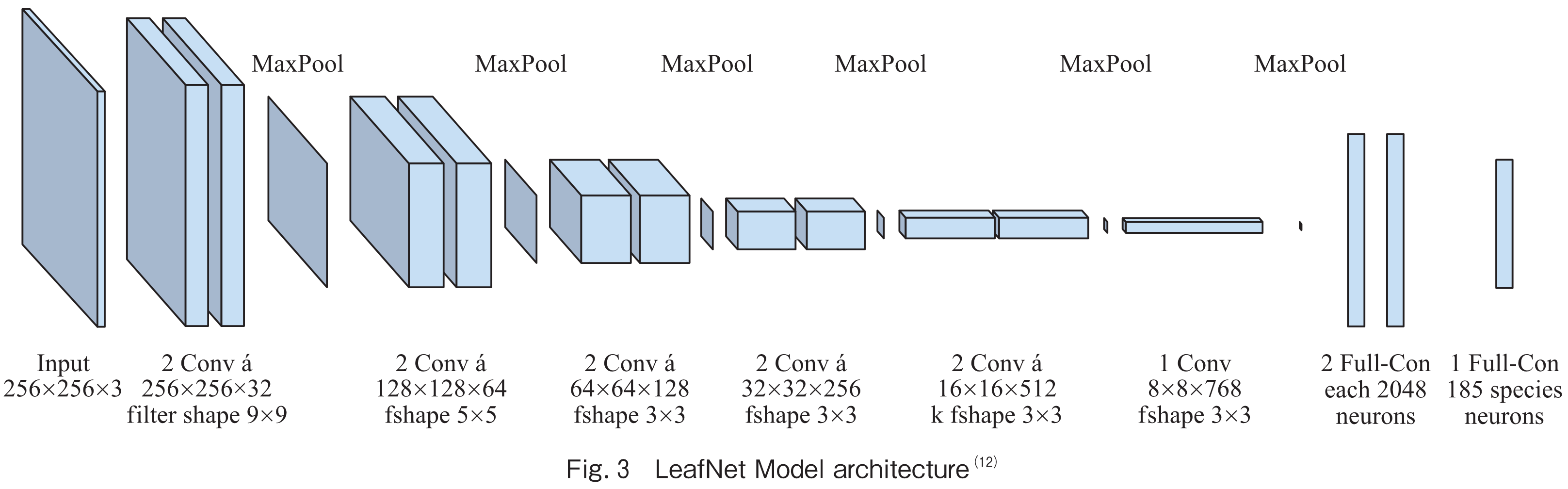
LeafNet is a CNN architecture proposed by Pierre Barré et. al.(12). Fig.3 illustrates the original architecture of the LeafNet model. The model architecture of LeafNet consists of three convolutional layers, three max-pooling layers, and two fully connected layers.
There are 64 filters in the first convolutional layer with a kernel size of 5×5, 128 filters in the second convolutional layer with a kernel size of 3×3, and 256 filters in the third convolutional layer with a kernel size of 3×3. The three convolutional layers are followed by one max-pooling layer each. The number of units in the 1,024 and 38 completely connected layers, respectively, corresponds to the number of leaf disease classes.
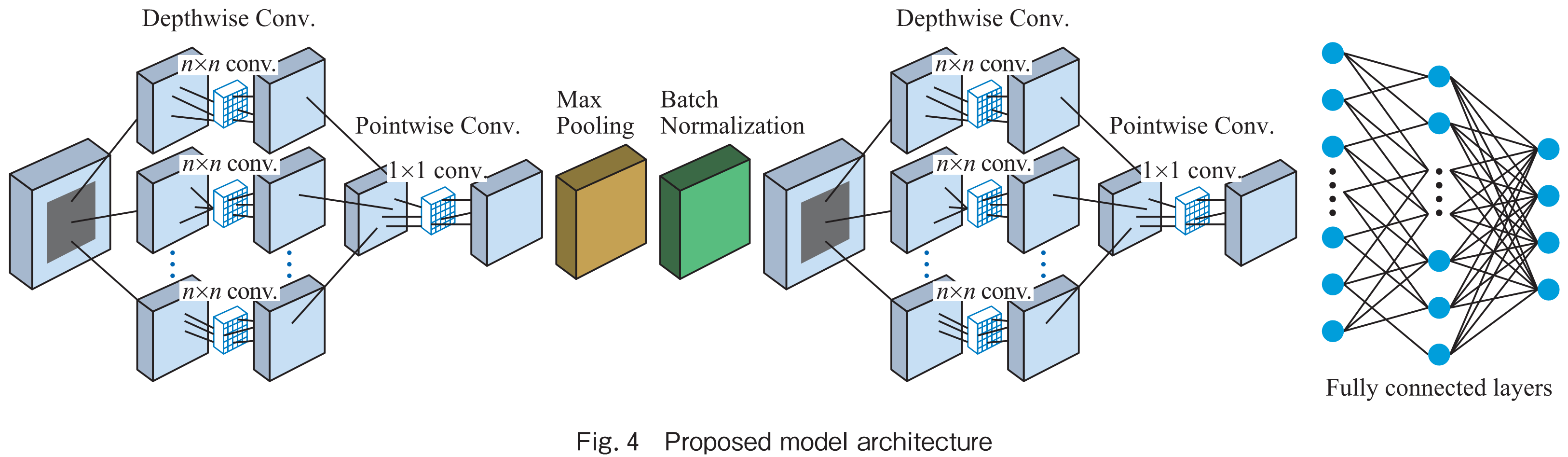
CNNs has been widely used architecture for image classification tasks. Depthwise separable convolution is a modification of the standard convolutional layer that reduces the number of computations while maintaining the accuracy of the model. This is achieved by splitting the convolution into two separate operations: a depthwise convolution and a pointwise convolution. In the depthwise convolution layer, each filter is applied to a single input channel, resulting in a set of output channels equal to the number of input channels. This process is followed by a pointwise convolution layer, where a 1×1 filter is applied to combine the output channels from the depthwise convolution. This process reduces the number of parameters required by the model and also reduces the computational cost of the convolution operation. The Depthwise Separable Convolution can be used as a replacement for the standard convolutional layer in a CNN architecture, allowing for the creation of more lightweight models that can be trained and executed on resource-constrained devices. The modified model used in this study is an adaptation of the original LeafNet model. The main modifications were the addition of depthwise and separable convolutional layers, which were inserted after the initial convolutional layers of the LeafNet architecture. These layers were used to extract more refined and diverse features from the input images. Specifically, the depthwise convolutional layers perform spatial convolutions on each input channel separately, while the separable convolutional layers combine depthwise and pointwise (1×1) convolutions to reduce the computational cost and improve the model’s efficiency. Fig.4 illustrates the architecture of the proposed model. The modified model architecture consists of four main blocks: (1) convolutional layers, (2) depthwise convolutional layers, (3) separable convolutional layers, and (4) fully connected layers. The convolutional layers have 32, 64, and 128 filters with a kernel size of 3×3, followed by a max-pooling layer with a stride of 2×2. The depthwise convolutional layers have 32, 64, and 128 filters with a kernel size of 3×3. The separable convolutional layers have 32 and 64 filters with a kernel size of 3×3. The fully connected layers consist of two layers with 512 and 38 units, respectively, followed by a softmax activation function to produce the final classification output.
The hyperparameters used in this study were carefully selected to optimize the performance of the proposed model. Three different optimizer e.g., Adam, RMSprop and SGD was used with a learning rate of 0.0001, which is a commonly used optimizer in deep learning. A batch size of 128 was chosen. The number of epochs was set to 50. The proposed model architecture contains a total of 12,134,086 parameters, out of which 12,133,126 are trainable parameters and 960 are non-trainable parameters. The proposed model architecture and hyperparameters were carefully chosen by random search method to optimize the performance while keeping the model lightweight. This ensures that the model can be trained and deployed efficiently on devices with limited computational resources. Table 1 given below described the achieved results on the basis of accuracies with three different optimizers. This table provides the evaluation metrics for three different optimizers (Adam, RMSprop, and SGD) on two different models (LeafNet and Proposed Model) for a classification task. The columns “Train-Acc” and “Valid-Acc” indicate the accuracy of the models on the training and validation sets, respectively. The higher the values, the better the model’s performance. The columns “Train-Loss” and “Valid-Loss” represent the loss of the models on the training and validation sets, respectively.
Evaluation metrics are crucial to assess the performance of a classification model. Precision, recall, and F1 score are important evaluation metrics that are calculated from the confusion matrix. Precision is defined as the ratio of true positives to the sum of true positives and false positives. It measures how many of the samples classified as positive are actually positive. Recall, on the other hand, is defined as the ratio of true positives to the sum of true positives and false negatives. It measures how many of the actual positive samples were correctly identified by the model. F1 score is the harmonic mean of precision and recall, and it provides a balanced measure of the model’s performance. These evaluation metrics provide a comprehensive understanding of the performance of the proposed model. The obtained results of precision, recall, F1 score and test accuracy has been depicted in Fig.5. Test accuracy represents the proportion of correct predictions made by the model out of all predictions.
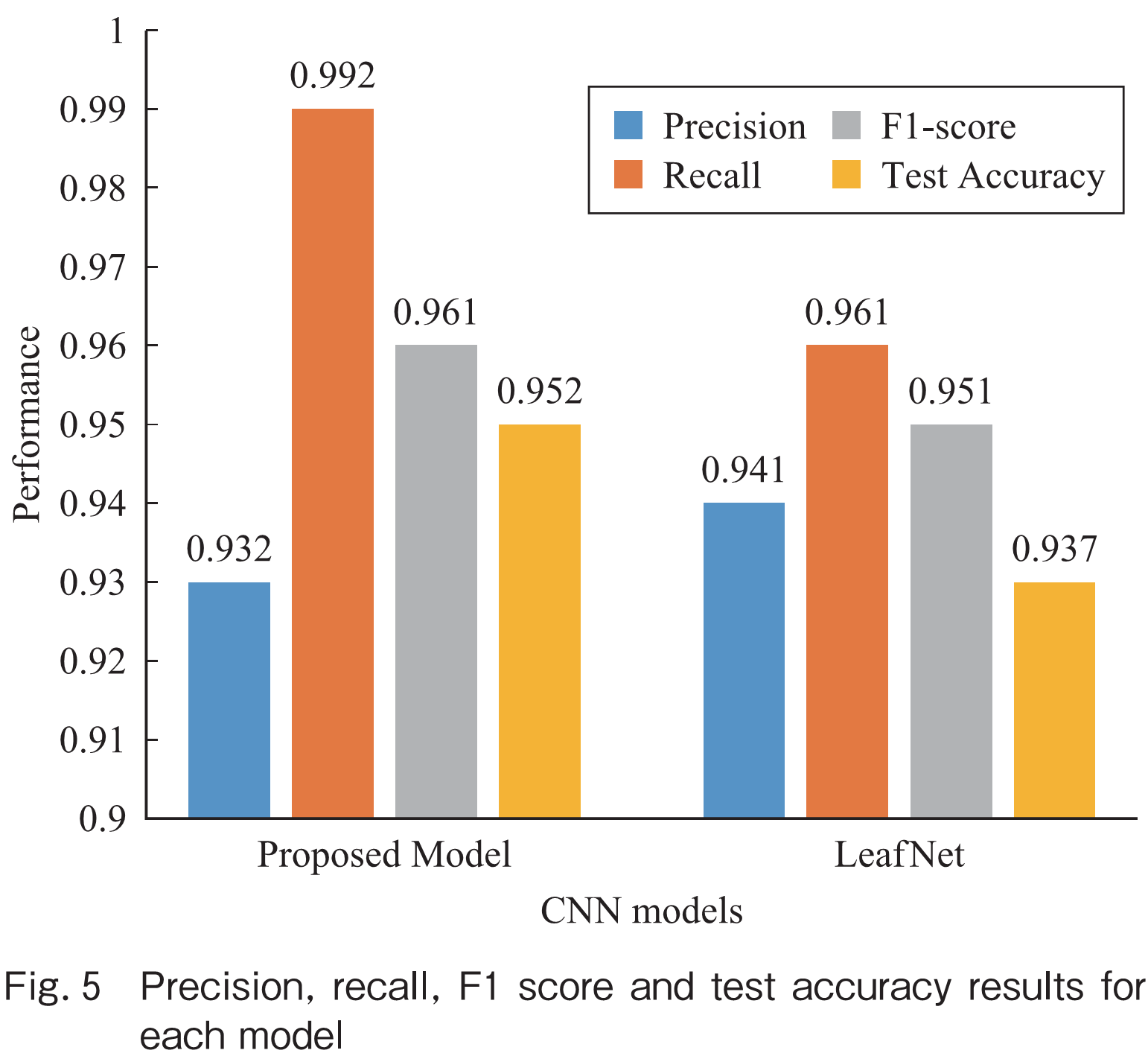
Looking at the results, it is evident that the Proposed Model outperformed LeafNet in all aspects except for precision. The Proposed Model achieved a test accuracy of 95.25%, which is significantly higher than the test accuracy of LeafNet at 93.76%. Moreover, the Proposed Model had a lower test loss of 0.181 than LeafNet model i.e., 0.213, indicating that it generalizes better to unseen data. These results suggest that the Proposed Model is a better choice for this particular task. When comparing the performance of the three optimizers, Adam consistently outperformed RMSprop and SGD. This is evident from the fact that the proposed model achieved the highest test accuracy and lowest test loss when trained using Adam. In contrast, the performance of RMSprop and SGD was lower across all models. Therefore, we can conclude that Adam is the optimal optimizer for this dataset. In this case, the proposed model outperformed LeafNet in all other aspects, indicating that it is a more robust and reliable model. According to the Fig.5, the proposed model has a higher recall value i.e., 0.992 compared to LeafNet with recall of 0.961. In addition, the F1-score is higher for the proposed model i.e., 0.961 as compared to LeafNet 0.951. The test accuracy for the proposed model is also higher i.e., 0.952 or 95.25% as compared to LeafNet i.e., 0.937 or 93.76%. Overall, the proposed model performs better than LeafNet based on these evaluation metrics.
The experimental results of our study suggest that the proposed model outperforms LeafNet in terms of accuracy, precision, recall, and F1-score. The proposed model achieved a test accuracy of 95.25%, which is a significant improvement over LeafNet accuracy of 93.76%. Additionally, the proposed model showed higher precision, recall, and F1-score than LeafNet. The results also indicate that the choice of optimizer has a significant impact on the performance of both LeafNet and the proposed model. The Adam optimizer showed the best performance for both models, followed by RMSprop and SGD. It is important to note that the proposed model achieved better performance with all three optimizers compared to LeafNet. Overall, the results of our study suggest that the proposed model has the potential to be a valuable tool in the classification of images. Further studies could investigate the generalizability and robustness of the proposed model to different types of data and environments.
Acknowledgments The authors would like to acknowledge the support of the Fundamental Research Grant Scheme (FRGS), FRGS/1/2021/ICT08/MMU/03/1, Ministry of Higher Education, Malaysia, and the Multimedia University IR Fund MMUI/220058.
(1) A.A. Hamad, “Utility of Cilefa Pink B, a food dye in a facile decoration of the first green molecular-size-based fluorescence probe (MSBFP) for determining trimebutine; application to bulk, dosage forms, and real plasma,” Spectrochim. Acta A: Mol. Biomol. Spectrosc., vol.288, p.122187, 2023.
(2) I. Roussaki, K. Doolin, A. Skarmeta, G. Routis, J. Antonio Lopez-Morales, E. Claffey, M. Mora, and J. Antonio Martinez, “Building an interoperable space for smart agriculture,” Digital Communications Networks, vol.9, no.1, pp.183-193, 2023.
(3) C.C. Okolie, G. Danso-Abbeam, O. Groupson-Paul, and A.A. Ogundeji, “Climate-smart agriculture amidst climate change to enhance agricultural production: A bibliometric analysis,” Land, vol.12, no.1, p.50, 2023.
(4) D. Pal and S. Joshi, “AI, IoT and robotics in smart farming: Current applications and future potentials,” 2023 International Conference on Sustainable Computing and Data Communication Systems (ICSCDS), 2023, pp.1096-1101, 2023.
(5) M. Javaid, A. Haleem, I.H. Khan, and R. Suman, “Understanding the potential applications of artificial intelligence in agriculture sector,” Advanced Agrochem, vol.2, no.1, pp.15-30, 2023.
(6) N.T.T. Nguyen, L.M. Nguyen, T.T.T. Nguyen, D.H. Nguyen, D.T.C. Nguyen, and T. Van Tran, “Recent advances on biogenic nanoparticles for detection and control of plant pathogens in sustainable agriculture: A review,” Industrial Crops Products, vol.198, p.116700, 2023.
(7) S. Kuyah, S. Buleti, K. Dimobe, L. Nkurunziza, S. Moussa, C. Muthuri, and I. Öborn, “Farmer-managed natural regeneration in Africa: Evidence for climate change mitigation and adaptation in drylands,” in Agroforestry for Sustainable Intensification of Agriculture in Asia and Africa, pp.53-88, Springer, 2023.
(8) M.P.J. Mahenge, H. Mkwazu, R.R. Madege, B. Mwaipopo, and C. Maro, “Artificial intelligence and deep learning based technologies for emerging disease recognition and pest prediction in beans (phaseolus vulgaris l.): A systematic review,” 2023.
(9) G. Edison, “Role of artificial intelligence in agriculture sector,” BULLET: Jurnal Multidisiplin Ilmu, vol.2, no.2, pp.272-276, 2023.
(10) M. Hofman-Bergholm, “A transition towards a food and agricultural system that includes both food security and planetary health,” Foods, vol.12, no.1, p.12, 2023.
(11) N.B.K. Zaman, W.N.A.A. Raof, A.R. Saili, N.N. Aziz, F.A. Fatah, and S.K. Vaiappuri, “Adoption of smart farming technology among rice farmers,” Journal of Advanced Research in Applied Sciences Engineering Technology, vol.29, no.2, pp.268-275, 2023.
(12) P. Barré, B.C. Stöver, K.F. Müller, and V. Steinhage, “LeafNet: A computer vision system for automatic plant species identification,” Ecological Informatics, vol.40, pp.50-56, 2017.
(13) A.G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang, T. Weyand, M. Andreetto, and H. Adam, “Mobilenets: Efficient convolutional neural networks for mobile vision applications,” arXiv preprint arXiv: .04861, 2017.
(14) A. Sharma, A. Jain, P. Gupta, and V. Chowdary, “Machine learning applications for precision agriculture: A comprehensive review,” IEEE Access, vol.9, pp.4843-4873, 2020.
(15) M. Umair, M.S. Khan, F. Ahmed, F. Baothman, F. Alqahtani, M. Alian, and J. Ahmad, “Detection of COVID-19 using transfer learning and grad-CAM visualization on indigenously collected X-ray dataset,” Sensors, vol.21, no.17, p.5813, 2021.
(16) H. Ahmed, M. Umair, A. Iftikhar, and K. Sultana, “Covid-19 variants detection & classification using self proposed two stage MNN-2: robust comparison with Yolo V5 & Faster R-CNN,” 2022 IEEE International Conference on Blockchain, Smart Healthcare and Emerging Technologies (SmartBlock4Health), pp.1-7, 2022.
(17) B.V. Gokulnath and U.D. Gandhi, “Identifying and classifying plant disease using resilient LF-CNN,” Ecological Informatics, vol.63, p.101283, 2021.
(18) J. Chen, D. Zhang, Y.A. Nanehkaran, and D. Li, “Detection of rice plant diseases based on deep transfer learning,” Journal of the Science of Food and Agriculture, vol.100, no.7, pp.3246-3256, 2020.
(19) J. Shijie, J. Peiyi, H. Siping, and s. Haibo, “Automatic detection of tomato diseases and pests based on leaf images,” 2017 Chinese Automation Congress (CAC), pp.2537-2510, 2017.
(20) R.P. Singh, P. Sahu, A. Chug, A. Prakash Singh, and D. Singh, “Deep learning models for beans crop diseases: Classification and visualization techniques,” International Journal of Modern Agriculture, vol.10, no.1, pp.796-812, 2021.
(21) V. Tiwari, R.C. Joshi, and M.K. Dutta, “Dense convolutional neural networks based multiclass plant disease detection and classification using leaf images,” Ecological Informatics, vol.63, pp.101289, 2021.
(22) S. Bhattarai, “New Plant Diseases Dataset.”
https://www.kaggle.com/datasets/vipoooool/new-plant-diseases-dataset
(Manuscript received May 31, 2023. Manuscript final received July 13, 2023.)
オープンアクセス以外の記事を読みたい方は、以下のリンクより電子情報通信学会の学会誌の購読もしくは学会に入会登録することで読めるようになります。 また、会員になると豊富な豪華特典が付いてきます。
電子情報通信学会 - IEICE会誌はモバイルでお読みいただけます。
電子情報通信学会 - IEICE会誌アプリをダウンロード